This task aims at segmentation of whole body disease burden on PSMA PET/CT (a mix of 68Ga-PSMA-617 and 18F-DCFPyL) and FDG PET/CT images for staging of patient for 177Lu-PSMA Therapy.
The training dataset involves 100 patients staged prior to LuPSMA therapy with both PSMA and FDG PET/CT. The data is provided in a format which allows evaluation of both imaging modalities for potentially improved segmentation by combined features in both study sets. The expected output annotations are independent total tumour burden (TTB) segmentations for each of the PSMA and FDG studies. Per case, the extent of disease will need to be determined for each tracer. This can be considered for each scan independently (eg PSMA- and FDG-only algorithms) or may attempt to utilise composite information from both scans may assist with determination of otherwise equivocal lesions when evaluated as a single modality.
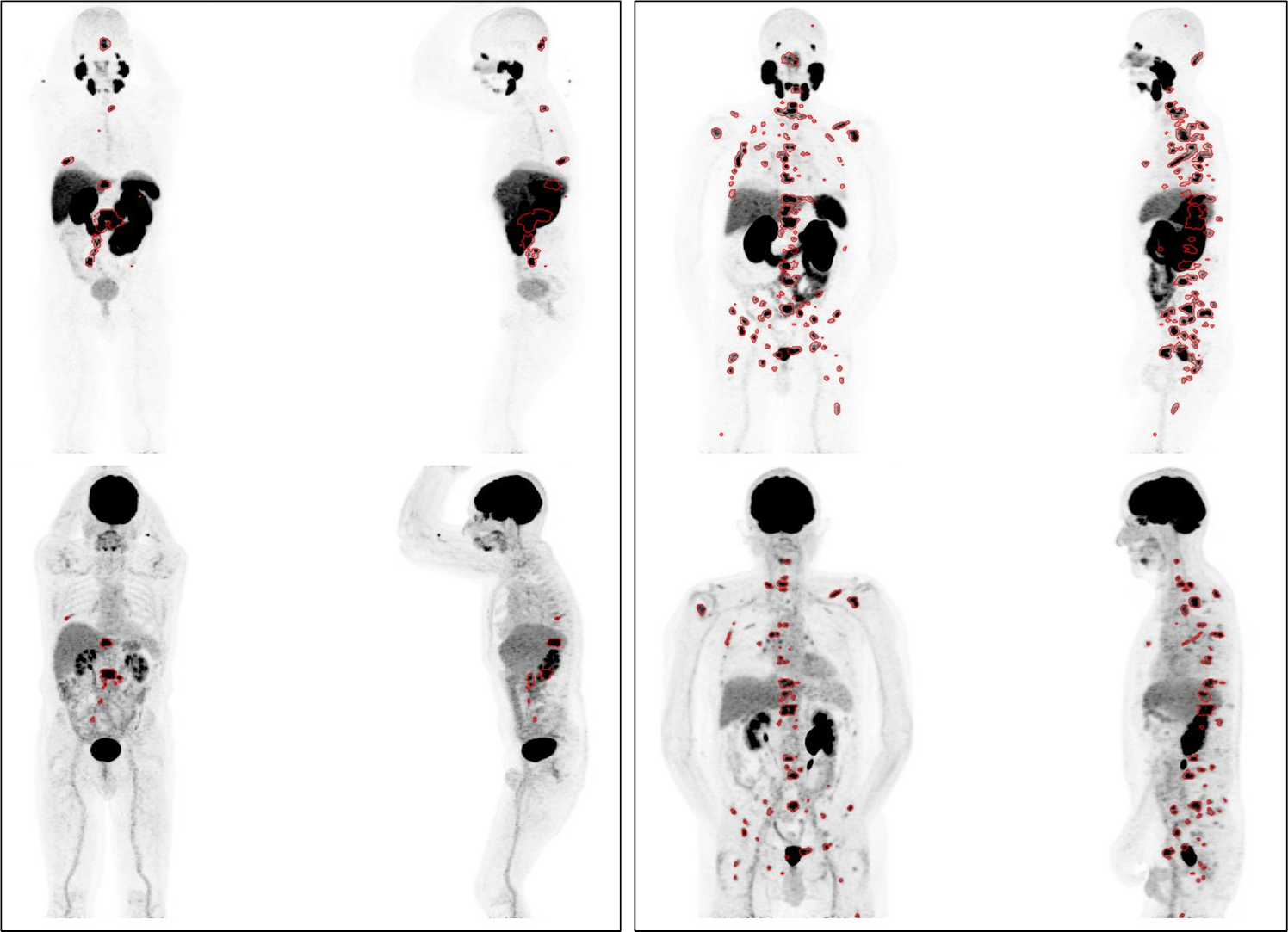 Example of annotated MIP images for two cases (left and right box). For each case, PSMA PET illustrated on top row and accompanying FDG PET in bottom row.
Example of annotated MIP images for two cases (left and right box). For each case, PSMA PET illustrated on top row and accompanying FDG PET in bottom row.
The images are provided in the native CT and PET resolutions with PET image data re-scaled to units of SUV (body weight). We provide the output of a course rigid registration to facilitate alignment between PSMA and FDG image sets and the predicted organ/bone/tissue segmentation from Total Segmentator (v2.4, task=total). We ask the No other additional pre-trained models or image data are used in this challenge.
Images are contoured based on a fixed SUV threshold per scan. All PSMA PET/CTs utilise SUV>=3 while FDG PET/CT images use a liver-based threshold (generally Liver Mean + 2*SD). The designated threshold for each study is provided with the image data. Subsequently, determination of malignant versus physiological areas of tracer avidity has been manually annotated by expert nuclear medicine physician with 5 or more years specialisation. All TTB labels are matched to the resolution of the PET image which is generally of slightly lower resolution than the CT component.
All cases will have some presence of disease for contouring but there will be a distribution of large- and small-volume cases and various patterns of metastatic spread (eg largely osseous, nodal, liver-involvement, etc). We have considered a balance of training and test cases that will account for a similar distribution of case patterns based on manual selection.
A live leaderboard will be available for benchmarking on a small validation dataset (10 cases) and subsequently the final evauluation will be based on a single, final submission on a 40-case test dataset. Best performing models will be invited to participate in the DEEP-PSMA workshop session at MICCAI 2025. We have taken care to include a selection of different cancer distributions, including known areas of difficult classification, in each of the three datasets (training, preliminary testing, final validation).
Data Acquisition: Image data acquired on standard diagnostic PET/CT systems. Predominantly from institutional scanners GE Discovery 710 & 690 PET/CTs, Siemens Biograph PET/CT, and Siemens Vision 600 PET/CT. Whole body (predominantly vertex to thighs) PET images with low-dose CT component for attenuation correction and anatomical localization. PET images reconstructed with standard corrections EANM EARL-compliant resolution recovery settings.
