All challenge submissions should be made by Docker container and will be run on the Grand Challenge hosting platform.
There is a 30 minute wall time limit per case which includes the time to contour one patient's PSMA and FDG PET/CT scans as well as any pre/post-processing operations. Essentially, the image, organ segmentation, and rigid co-registration parameters shown in the Dataset Format page will be visible to the algorithm with the expectation to generate the Total Tumour Burden (TTB) segmentation for FDG and PET modalities. Deployment will be able to utilise Nvidia T4 GPUs (ml.g4dn instances) as shown in the runtime environment documentation.
If you have previously participated in a recent Grand Challenge, the format for submission should be very similar to previous competitions. General documentation is provided here however it may be easiest to adapt the baseline algorithm from our Github page. You will need a docker installation on your PC and if using windows it may be necessary to install WSL (windows subsystem linux) with Docker to ensure that the container is compatible with the deployment environment.
Example Docker Container - Github
For simplicity, the example algorithm and docker workflows are shown based on creating a single tar.gz image encompassing the user's runtime code and model weights however there are more advanced functionalities on the Grand Challenge platform that allow updating just portions of the model files and weights to more easily update algorithms and save re-uploading an entire image (often 10+ GB).
Submitting an Algorithm
Click the submit button below the banner and select the relevant phase to apply (Debug, Preliminary Testing, Final Validation). In the first instance, you will need to upload the tar of your docker container image with the + Manage your algorithms button on the right of the page (about halfway down). On this page you can maintain up to 3 live algorithms for the challenge. Once exceeded, you will have to update one of the slots with any updates. For each algorithm you can configure the base docker tar image, any supplementary model .tar file and the details of the required runtime environment (eg T4 GPU 16 GB VRAM, 32 GB System RAM). For the Debug phase, this should also be the location that you can review the debug output (Stdout/Stderr). From the algorithm page, select Results, click the [i] info icon next to a case, and on the page select >_Logs in the upper left corner. You should also be able to review the output segmentation on the Open Results Viewer button or by downloading either of the output .mha images.
Once your algorithm is configured. Return to the Create New Submission page and it should now be visible to select in the Algorithm* dropdown page. Click save and the system will begin to queue the jobs and there will be a notification once completed.
Leaderboards
For the Debug and Preliminary Validation Phases, the leaderboard position will be automatically updated based on the most recent completed submission with working result evaluation. This may be used for fine-tuning your algorithms based on the 10 unseen cases in the preliminary validation phase and selecting which model as your last submission in the final testing phase. For the final phase (40 cases), the leaderboard position will be based on the last working result with a limit of 3 submissions per participant. The leaderboard standing for the final testing phase will be released in the week following the challenge close date on 19 August. The final standings will only include submissions providing a methodological paper submission and link to training source code.
Increasing System DRAM to 32 GB
The VRAM is limited to 16 GB and the system DRAM defaults to 16 in the config page and can be increased to 32 in the text box configurations
This is how to update for an existing model and resubmit.
 .
.
To access your algorithm configurations. Click to submit to one of the phases.
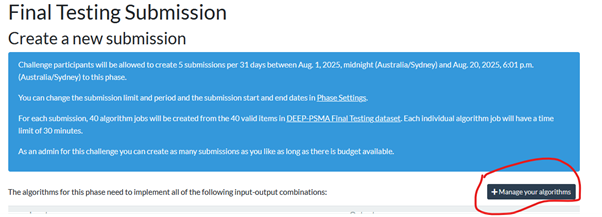
Select “manage your algorithms”
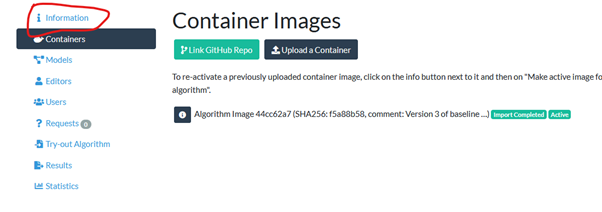
To edit the settings of your algorithm, click “manage container images for” the one to adjust

Select the “information” tab and click “update settings”
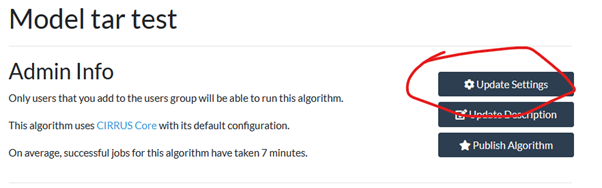
About halfway down the page there is the setting for system DRAM which defaults to 16 but can be entered up to 32.
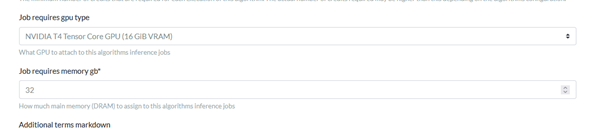
Click save and the setting will be updated.
The platform won’t let you resubmit an identical docker algorithm+model tar combination to a phase that you’ve already lodged a job for. That’s even if adjusting the model configuration.
If you haven’t utilised a separate model.tar file to include your weights at runtime, the easiest thing is to create any small .tar file which can be uploaded and allow re-running the container. Otherwise, you’ll need to edit the main algorithm container or model .tar files such that one of them has an updated SHA hash (eg add a print statement), re-upload the container/model image and it will permit re-running on a phase with an existing submission.
