General Tips to Improve Model Performance¶
For this challenge, it's important to take stock of the manual contouring methodology used to generate the ground truth contours. For delineating tumour boundary, the physicians have utilised the methods reported in the manuscripts by Buteau et al. and Ferdinandus et al. which are designed for highly reproducible scoring of PET biomarkers.
The outline of most contour surfaces will be based on the threshold boundary for the respective PET image modality: PSMA SUV>3, and FDG Liver-based. The appropriate values to use for each case are provided in the training data and are accessible as objects in the grand challenge evaluation docker environment for test cases. See Lines 78 and 90 in our example inference script and docker container for how to access the threshold values at runtime.
A simple way to take advantage of the threshold methodology to improve the performance of a deep learning model is to apply a post-processing refinement that adjusts the boundary of the initial inference to ensure that no voxels below the threshold are included in the final segmentation and provides scope to expand the boundary to the threshold level for lesions that are initially smaller than the ground truth annotation.
Our Baseline Algorithm includes this in some of the processing steps after initial segmentation with an optimised nnU-Net model. Alternatively, we have provided a one-step function which provides the same functionality with the additional scope to omit label expansion into certain areas of known physiological uptake such as liver (total segmentator label # 5).
Please consider this as a refinement method for your submitted algorithms!¶
This offers a very simple, but significant improvement for the DEEP-PSMA task. As an example, for the validation cases in Fold 0 of our Baseline PSMA PET model this improves the average TTB Dice score from 0.867 to 0.933.
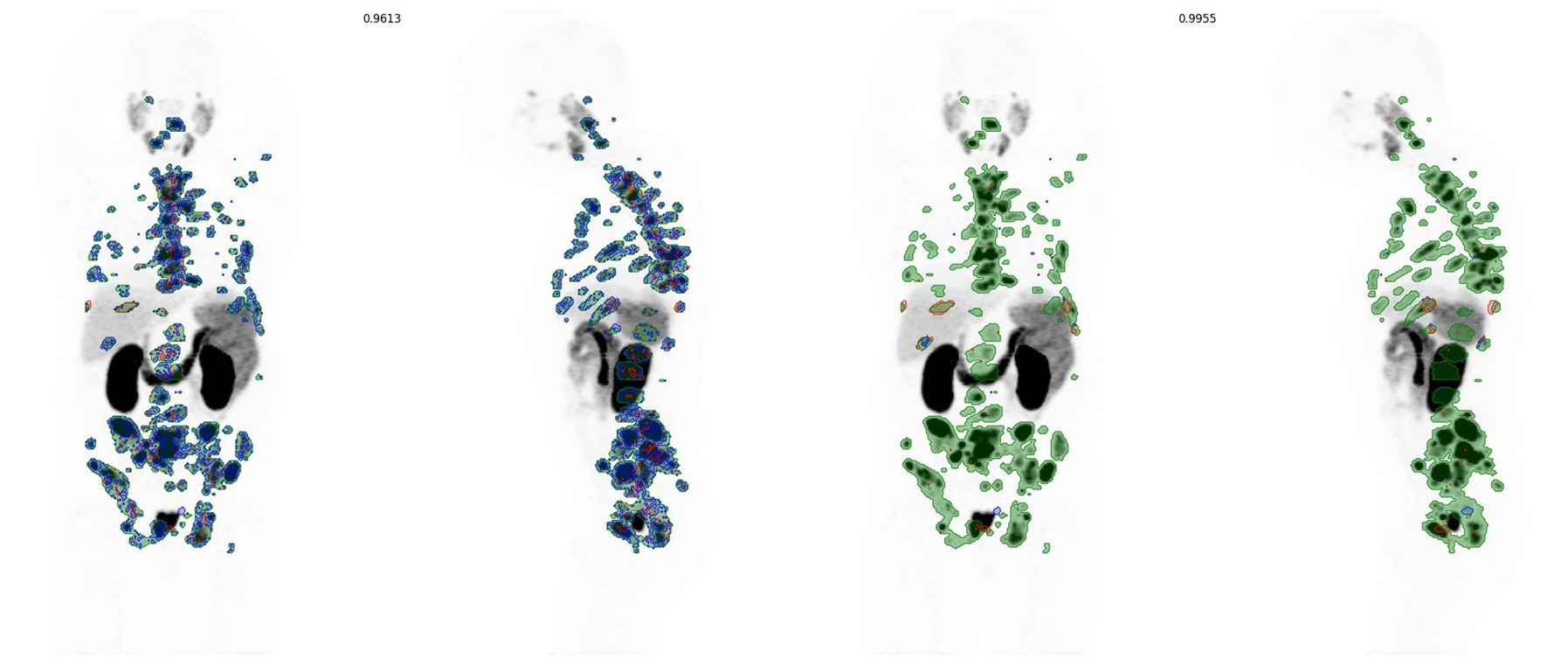 Initial validation output for case train_0005 (left, dice 0.961) and refined segmentation with expansion and re-thresholding (rightm, dice 0.996). Areas of green are good alignment with ground truth and prediction while false negative voxels are shown in blue and false positive regions in red.
Initial validation output for case train_0005 (left, dice 0.961) and refined segmentation with expansion and re-thresholding (rightm, dice 0.996). Areas of green are good alignment with ground truth and prediction while false negative voxels are shown in blue and false positive regions in red.
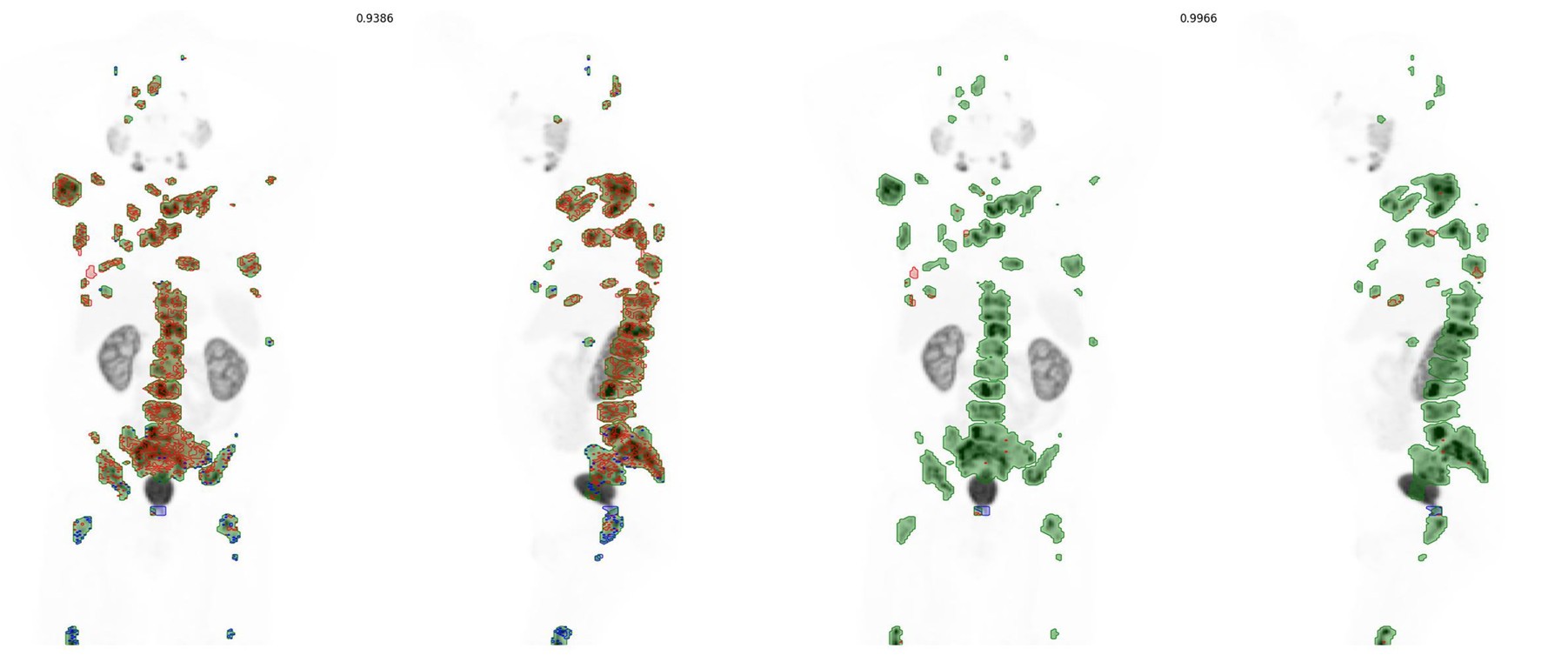 Original and refined contouring for case train_0014. Initially inferred label includes many areas extending beyond the appropriate SUV threshold and a few regions in the femurs that need to be expanded. After refinement, most of the surface is in close agreement and dice score improves from 0.939 to 0.997.
Original and refined contouring for case train_0014. Initially inferred label includes many areas extending beyond the appropriate SUV threshold and a few regions in the femurs that need to be expanded. After refinement, most of the surface is in close agreement and dice score improves from 0.939 to 0.997.
Again please consider these steps in your models as simple ways to quite reliably improve performance for the task in this Grand Challenge. One more link to the refinement code. There are options to adjust the initial TTB growth distance and which anatomical VOIs are ignored for refinement. We leave it to you to experiment with some of the potential parameters. From quick tests, adjusting the initial expansion radius between 5-10 mm did not appreciably alter the final label accuracy and omitting the expansion in the vicinity of liver for PSMA PET is advisable.
